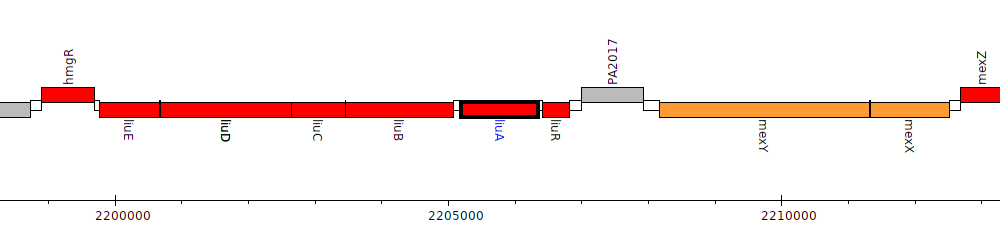
Pseudomonas aeruginosa PAO1, PA2015 (liuA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0008470 | isovaleryl-CoA dehydrogenase activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
18625020 | Reviewed by curator |
| Molecular Function | GO:0008470 | isovaleryl-CoA dehydrogenase activity | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
16517656 | Reviewed by curator |
| Biological Process | GO:0008300 | isoprenoid catabolic process | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
16517656 | Reviewed by curator |
| Biological Process | GO:0044248 | cellular catabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0003995 | acyl-CoA dehydrogenase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd01156
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0050660 | flavin adenine dinucleotide binding |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:1.10.540.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:1.10.540.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae00280 | Valine, leucine and isoleucine degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Acyclic isoprenoid catabolism |
ECO:0000037
not_recorded |
|||
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF02770 | Acyl-CoA dehydrogenase, middle domain | IPR006091 | Acyl-CoA oxidase/dehydrogenase, middle domain | 127 | 222 | 7.8E-27 |
| Pfam | PF00441 | Acyl-CoA dehydrogenase, C-terminal domain | IPR009075 | Acyl-CoA dehydrogenase/oxidase, C-terminal | 234 | 382 | 4.1E-41 |
| PIRSF | PIRSF016578 | PIGM | - | - | 12 | 387 | 1.2E-14 |
| FunFam | G3DSA:2.40.110.10:FF:000004 | Isovaleryl-CoA dehydrogenase, mitochondrial | - | - | 126 | 239 | 4.0E-57 |
| SUPERFAMILY | SSF47203 | Acyl-CoA dehydrogenase C-terminal domain-like | IPR036250 | Acyl-CoA dehydrogenase-like, C-terminal | 233 | 385 | 6.28E-46 |
| FunFam | G3DSA:1.10.540.10:FF:000022 | Isovaleryl-CoA dehydrogenase isoform 2 | - | - | 2 | 124 | 8.4E-58 |
| Gene3D | G3DSA:2.40.110.10 | - | IPR046373 | Acyl-CoA oxidase/dehydrogenase, middle domain superfamily | 126 | 239 | 5.0E-43 |
| Gene3D | G3DSA:1.10.540.10 | - | IPR037069 | Acyl-CoA dehydrogenase/oxidase, N-terminal domain superfamily | 2 | 124 | 6.0E-40 |
| Pfam | PF02771 | Acyl-CoA dehydrogenase, N-terminal domain | IPR013786 | Acyl-CoA dehydrogenase/oxidase, N-terminal | 14 | 123 | 2.3E-38 |
| FunFam | G3DSA:1.20.140.10:FF:000003 | isovaleryl-CoA dehydrogenase, mitochondrial | - | - | 240 | 384 | 3.7E-80 |
| SUPERFAMILY | SSF56645 | Acyl-CoA dehydrogenase NM domain-like | IPR009100 | Acyl-CoA dehydrogenase/oxidase, N-terminal and middle domain superfamily | 5 | 247 | 1.2E-86 |
| Gene3D | G3DSA:1.20.140.10 | - | - | - | 240 | 382 | 1.1E-46 |
| CDD | cd01156 | IVD | IPR034183 | Isovaleryl-CoA dehydrogenase | 9 | 384 | 0.0 |
| PANTHER | PTHR43884 | ACYL-COA DEHYDROGENASE | - | - | 12 | 385 | 0.0 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.