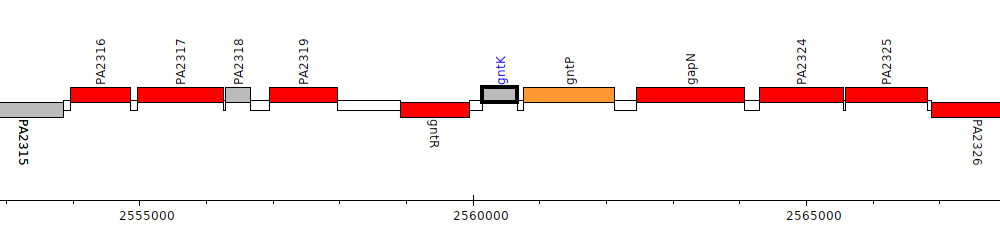
Pseudomonas aeruginosa PAO1, PA2321 (gntK)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0004340 | glucokinase activity | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
8655507 | Reviewed by curator |
| Biological Process | GO:0015976 | carbon utilization | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0006091 | generation of precursor metabolites and energy | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0044248 | cellular catabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0005975 | carbohydrate metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR43442
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016301 | kinase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR43442
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Glycolysis / Gluconeogenesis |
ECO:0000037
not_recorded |
|||
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01120 | Microbial metabolism in diverse environments | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00030 | Pentose phosphate pathway | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | GLUCONSUPER-PWY | D-gluconate degradation | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCyc | IDNCAT-PWY | L-idonate degradation | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01200 | Carbon metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Pentose phosphate cycle |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 5 | 170 | 1.1E-52 |
| CDD | cd02021 | GntK | IPR006001 | Carbohydrate kinase, thermoresistant glucokinase | 8 | 152 | 1.91281E-63 |
| FunFam | G3DSA:3.40.50.300:FF:000522 | Gluconokinase | - | - | 3 | 170 | 2.5E-61 |
| Pfam | PF13671 | AAA domain | - | - | 9 | 139 | 2.5E-11 |
| PANTHER | PTHR43442 | GLUCONOKINASE-RELATED | IPR006001 | Carbohydrate kinase, thermoresistant glucokinase | 7 | 157 | 1.2E-41 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 7 | 167 | 1.75E-25 |
| NCBIfam | TIGR01313 | JCVI: gluconokinase, GntK/IdnK-type | IPR006001 | Carbohydrate kinase, thermoresistant glucokinase | 8 | 168 | 5.0E-65 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.