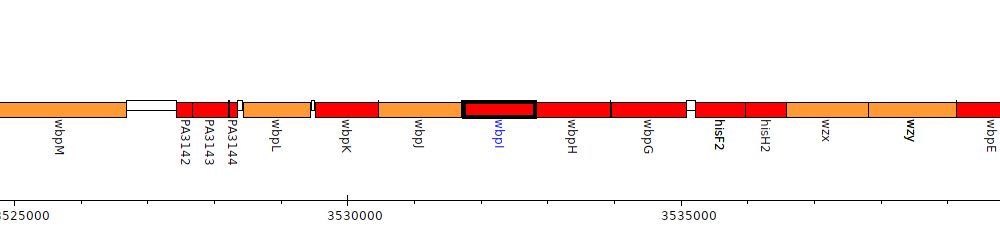
Pseudomonas aeruginosa PAO1, PA3148 (wbpI)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009243 | O antigen biosynthetic process | Inferred from Mutant Phenotype | ECO:0000315 mutant phenotype evidence used in manual assertion |
10930727 | Reviewed by curator |
| Molecular Function | GO:0050378 | UDP-glucuronate 4-epimerase activity | Inferred from Experiment | ECO:0000269 experimental evidence used in manual assertion |
17346239 | Reviewed by curator |
| Molecular Function | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
10930727 | Reviewed by curator |
| Biological Process | GO:0000271 | polysaccharide biosynthetic process | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
10930727 | Reviewed by curator |
| Molecular Function | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR43174
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Lipopolysaccharide biosynthesis |
ECO:0000037
not_recorded |
|||
| PseudoCAP | UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid biosynthesis pathway |
ECO:0000037
not_recorded |
|||
| KEGG | pae00520 | Amino sugar and nucleotide sugar metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF02350 | UDP-N-acetylglucosamine 2-epimerase | IPR003331 | UDP-N-acetylglucosamine 2-epimerase domain | 22 | 351 | 6.1E-111 |
| PANTHER | PTHR43174 | UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE | IPR029767 | UDP-N-acetylglucosamine 2-epimerase WecB-like | 1 | 353 | 8.2E-121 |
| NCBIfam | TIGR00236 | JCVI: UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing) | IPR029767 | UDP-N-acetylglucosamine 2-epimerase WecB-like | 1 | 352 | 1.7E-95 |
| Gene3D | G3DSA:3.40.50.2000 | Glycogen Phosphorylase B; | - | - | 1 | 192 | 1.9E-70 |
| SUPERFAMILY | SSF53756 | UDP-Glycosyltransferase/glycogen phosphorylase | - | - | 1 | 353 | 1.85E-105 |
| CDD | cd03786 | GTB_UDP-GlcNAc_2-Epimerase | - | - | 2 | 352 | 0.0 |
| Gene3D | G3DSA:3.40.50.2000 | Glycogen Phosphorylase B; | - | - | 193 | 333 | 1.1E-46 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.