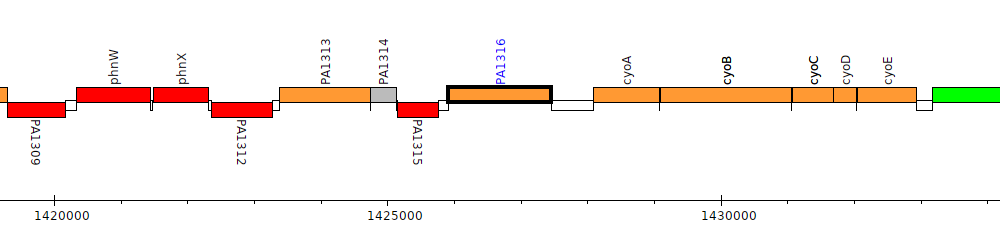
Pseudomonas aeruginosa PAO1, PA1316
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA1316
|
| Name |
|
| Replicon | chromosome |
| Genomic location | 1425912 - 1427453 (+ strand) |
| Transposon Mutants | 4 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 7 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_250007.1 |
| GI | 15596513 |
| Affymetrix | PA1316_at |
| Entrez | 881564 |
| GenBank | AAG04705.1 |
| INSDC | AAG04705.1 |
| NCBI Locus Tag | PA1316 |
| protein_id(GenBank) | gb|AAG04705.1|AE004561_1|gnl|PseudoCAP|PA1316 |
| TIGR | NTL03PA01317 |
| UniParc | UPI00000C530B |
| UniProtKB Acc | Q9I428 |
| UniProtKB ID | Q9I428_PSEAE |
| UniRef100 | UniRef100_A0A0C6EL89 |
| UniRef50 | UniRef50_G2E188 |
| UniRef90 | UniRef90_A6V8P4 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
probable major facilitator superfamily (MFS) transporter
|
| Synonyms |
probable MFS transporter |
| Evidence for Translation | |
| Charge (pH 7) | 6.55 |
| Kyte-Doolittle Hydrophobicity Value | 0.843 |
| Molecular Weight (kDa) | 53.1 |
| Isoelectric Point (pI) | 9.16 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 541 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG001213 (1391 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA1316
|
Human Homologs
References
|
Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen.
Lewenza S, Gardy JL, Brinkman FS, Hancock RE
Genome Res 2005 Feb;15(2):321-9
PubMed ID: 15687295
|
|
Nucleotide sequence analysis reveals similarities between proteins determining methylenomycin A resistance in Streptomyces and tetracycline resistance in eubacteria.
Neal RJ, Chater KF
Gene 1987;58(2-3):229-41
PubMed ID: 2828187
|
|
Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin.
Bibb MJ, Sherman DH, Omura S, Hopwood DA
Gene 1994 May 3;142(1):31-9
PubMed ID: 8181754
|
|
Cloning and sequence analysis of the putative rifamycin polyketide synthase gene cluster from Amycolatopsis mediterranei.
Schupp T, Toupet C, Engel N, Goff S
FEMS Microbiol Lett 1998 Feb 15;159(2):201-7
PubMed ID: 9503613
|