Pseudomonas aeruginosa PAO1, PA0706 (cat)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
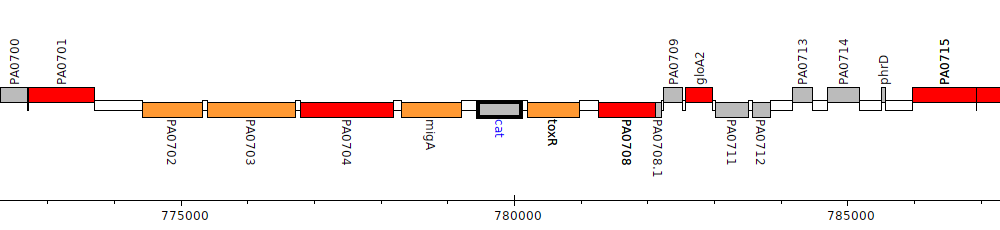
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA0706
|
| Name |
cat
|
| Replicon | chromosome |
| Genomic location | 779463 - 780101 (- strand) |
| Transposon Mutants | 2 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 1 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_249397.1 |
| GI | 15595903 |
| Affymetrix | PA0706_cat_at |
| DNASU | PaCD00006322 |
| Entrez | 878374 |
| GenBank | AAG04095.1 |
| INSDC | - |
| NCBI Locus Tag | PA0706 |
| protein_id(GenBank) | gb|AAG04095.1|AE004506_5|gnl|PseudoCAP|PA0706 |
| TIGR | NTL03PA00707 |
| UniParc | UPI000012703F |
| UniProtKB Acc | P26841 |
| UniProtKB ID | CAT4_PSEAE |
| UniRef100 | UniRef100_P26841 |
| UniRef50 | UniRef50_P26841 |
| UniRef90 | UniRef90_P26841 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
chloramphenicol acetyltransferase
|
| Synonyms |
xenobiotic acetyltransferase |
| Evidence for Translation | |
| Charge (pH 7) | -4.18 |
| Kyte-Doolittle Hydrophobicity Value | -0.196 |
| Molecular Weight (kDa) | 23.5 |
| Isoelectric Point (pI) | 6.07 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
AMR gene predictions from CARD database
Identified using the Resistance Gene Identifier (RGI)
| Model Name | Definition | Accession | Model Type (?) | SNP | Cutoff | Bit Score | Percent Identity | CARD Version |
| Pseudomonas aeruginosa catB7 | catB7 is a chromosome-encoded variant of the cat gene found in Pseudomonas aeruginosa. [PMID:10361706, PMID:10984043] | ARO:3002679 | protein homolog model | Perfect | 458.4 | 100.0 | 3.2.5 |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 2XAT | ACETYLTRANSFERASE | 03/11/98 | COMPLEX OF THE HEXAPEPTIDE XENOBIOTIC ACETYLTRANSFERASE WITH CHLORAMPHENICOL AND DESULFO-COENZYME A | Pseudomonas aeruginosa | 3.2 | X-RAY DIFFRACTION | 98.6 |
| 1XAT | ACETYLTRANSFERASE | 03/11/98 | STRUCTURE OF THE HEXAPEPTIDE XENOBIOTIC ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA | Pseudomonas aeruginosa | 3.2 | X-RAY DIFFRACTION | 98.6 |
Drugs Targeting this Protein
Identified by Diamond using e-value cutoff of 0.0001 and returning alignments that span 100% of the query sequence and that have more than 95% identity.
| Drug Name | Source Accession | Source DB | Version | Target Accession | Target Description | Percent Identity | Alignment Length | E-Value |
| Desulfo-Coenzyme A | DB01829 | DrugBank | 5.1.4 | P26841 | Chloramphenicol acetyltransferase | 100.0 | 212 | 6.0e-127 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 396 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000683 (866 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA0706
Search term: cat
Search term: chloramphenicol acetyltransferase
|
Human Homologs
References
|
Structure of the hexapeptide xenobiotic acetyltransferase from Pseudomonas aeruginosa.
Beaman TW, Sugantino M, Roderick SL
Biochemistry 1998 May 12;37(19):6689-96
PubMed ID: 9578552
|