Pseudomonas aeruginosa PAO1, PA4230 (pchB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
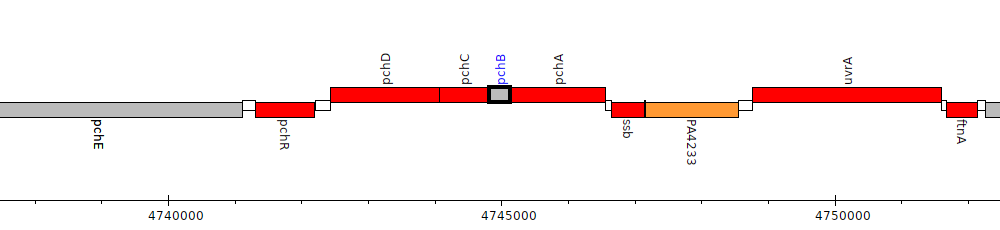
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA4230
|
| Name |
pchB
|
| Replicon | chromosome |
| Genomic location | 4744819 - 4745124 (+ strand) |
| Transposon Mutants | 1 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_252920.1 |
| GI | 15599426 |
| Affymetrix | PA4230_pchB_at |
| Entrez | 881846 |
| GenBank | AAG07618.1 |
| INSDC | AAG07618.1 |
| NCBI Locus Tag | PA4230 |
| protein_id(GenBank) | gb|AAG07618.1|AE004840_4|gnl|PseudoCAP|PA4230 |
| TIGR | NTL03PA04231 |
| UniParc | UPI00001313E9 |
| UniProtKB Acc | Q51507 |
| UniProtKB ID | PCHB_PSEAE |
| UniRef100 | UniRef100_Q51507 |
| UniRef50 | UniRef50_Q51507 |
| UniRef90 | UniRef90_Q51507 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
salicylate biosynthesis protein PchB
|
| Synonyms |
chorismate mutase isochorismate-pyruvate lyase |
| Evidence for Translation | |
| Charge (pH 7) | -1.80 |
| Kyte-Doolittle Hydrophobicity Value | -0.282 |
| Molecular Weight (kDa) | 11.4 |
| Isoelectric Point (pI) | 5.19 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 3REM | LYASE | 04/04/11 | Structure of the Isochorismate-Pyruvate Lyase from Pseudomonas aerugionsa with Bound Salicylate and Pyruvate | Pseudomonas aeruginosa | 1.95 | X-RAY DIFFRACTION | 100.0 |
| 2H9D | LYASE | 06/09/06 | Pyruvate-Bound Structure of the Isochorismate-Pyruvate Lyase from Pseudomonas aerugionsa | Pseudomonas aeruginosa | 1.95 | X-RAY DIFFRACTION | 100.0 |
| 3HGX | LYASE | 05/14/09 | Crystal Structure of Pseudomonas aeruginosa Isochorismate-Pyruvate Lyase K42A mutant in complex with salicylate and pyruvate | Pseudomonas aeruginosa | 2.5 | X-RAY DIFFRACTION | 99.0 |
| 3RET | LYASE | 04/05/11 | Salicylate and Pyruvate Bound Structure of the Isochorismate-Pyruvate Lyase K42E Mutant from Pseudomonas aerugionsa | Pseudomonas aeruginosa | 1.79 | X-RAY DIFFRACTION | 99.0 |
| 2H9C | LYASE | 06/09/06 | Native Crystal Structure of the Isochorismate-Pyruvate Lyase from Pseudomonas aeruginosa | Pseudomonas aeruginosa | 2.35 | X-RAY DIFFRACTION | 100.0 |
| 3HGW | LYASE | 05/14/09 | Apo Structure of Pseudomonas aeruginosa Isochorismate-Pyruvate Lyase I87T mutant | Pseudomonas aeruginosa | 2.25 | X-RAY DIFFRACTION | 99.0 |
Virulence Evidence
| Source 1 | |
| Database | VFDB : VF0095 |
| Category | Pyochelin |
| Evidence |
|
| Host Organism | |
| Notes | |
| Reference | |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 62 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000704 (672 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA4230
Search term: pchB
Search term: salicylate biosynthesis protein PchB
|
Human Homologs
References
No references are associated with this feature.