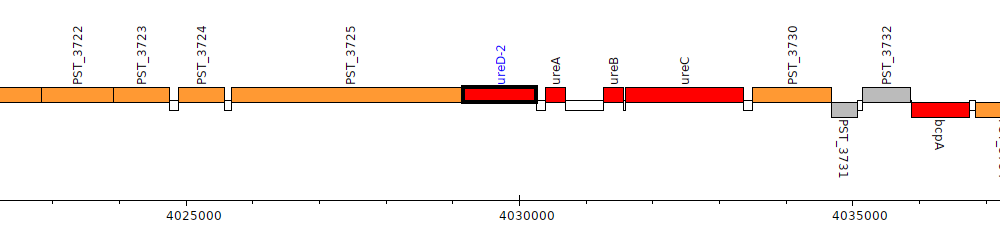
Pseudomonas stutzeri A1501, PST_3726 (ureD-2)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0016151 | nickel cation binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF01774
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006807 | nitrogen compound metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:PF01774
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Hamap | MF_01384 | Urease accessory protein UreD [ureD]. | IPR002669 | Urease accessory protein UreD | 97 | 346 | 29.399612 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 1 | 53 | - |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 22 | 39 | - |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 1 | 20 | - |
| PANTHER | PTHR33643 | UREASE ACCESSORY PROTEIN D | IPR002669 | Urease accessory protein UreD | 99 | 311 | 6.4E-30 |
| Pfam | PF01774 | UreD urease accessory protein | IPR002669 | Urease accessory protein UreD | 140 | 335 | 8.4E-49 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.