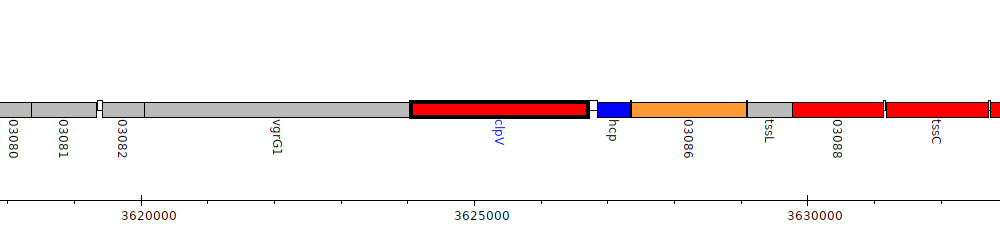
Pseudomonas sp. UW4, PputUW4_03084 (clpV)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0016887 | ATPase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF07724
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PR00300
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | ppuu03070 | Bacterial secretion system | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | ppuu02025 | Biofilm formation - Pseudomonas aeruginosa | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 605 | 623 | 1.4E-31 |
| Pfam | PF07724 | AAA domain (Cdc48 subfamily) | IPR003959 | ATPase, AAA-type, core | 601 | 765 | 1.4E-39 |
| Pfam | PF17871 | AAA lid domain | IPR041546 | ClpA/ClpB, AAA lid domain | 369 | 459 | 1.7E-29 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 189 | 506 | 1.79E-68 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 369 | 548 | 6.6E-39 |
| SMART | SM00382 | AAA_5 | IPR003593 | AAA+ ATPase domain | 223 | 368 | 1.9E-12 |
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 650 | 668 | 1.4E-31 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 550 | 847 | 2.7E-58 |
| Gene3D | G3DSA:1.10.8.60 | - | - | - | 772 | 868 | 2.6E-9 |
| Gene3D | G3DSA:1.10.1780.10 | - | IPR036628 | Clp, N-terminal domain superfamily | 2 | 160 | 5.1E-43 |
| Pfam | PF02861 | Clp amino terminal domain, pathogenicity island component | IPR004176 | Clp, repeat (R) domain | 23 | 66 | 0.0067 |
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 679 | 697 | 1.4E-31 |
| FunFam | G3DSA:3.40.50.300:FF:000025 | ATP-dependent Clp protease subunit | - | - | 554 | 770 | 4.5E-77 |
| FunFam | G3DSA:3.40.50.300:FF:000010 | Chaperone clpB 1, putative | - | - | 175 | 367 | 4.2E-83 |
| SMART | SM01086 | ClpB_D2_small_2 | IPR019489 | Clp ATPase, C-terminal | 772 | 865 | 7.0E-10 |
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 712 | 726 | 1.4E-31 |
| Pfam | PF00004 | ATPase family associated with various cellular activities (AAA) | IPR003959 | ATPase, AAA-type, core | 228 | 340 | 3.6E-10 |
| Coils | Coil | Coil | - | - | 135 | 155 | - |
| NCBIfam | TIGR03345 | JCVI: type VI secretion system ATPase TssH | IPR017729 | AAA+ ATPase ClpV1 | 12 | 850 | 0.0 |
| SUPERFAMILY | SSF81923 | Double Clp-N motif | IPR036628 | Clp, N-terminal domain superfamily | 12 | 127 | 3.53E-19 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 177 | 367 | 9.1E-71 |
| SMART | SM00382 | AAA_5 | IPR003593 | AAA+ ATPase domain | 601 | 751 | 7.3E-10 |
| CDD | cd19499 | RecA-like_ClpB_Hsp104-like | - | - | 562 | 731 | 6.26101E-75 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 554 | 769 | 3.1E-66 |
| Pfam | PF10431 | C-terminal, D2-small domain, of ClpB protein | IPR019489 | Clp ATPase, C-terminal | 773 | 850 | 1.2E-12 |
| CDD | cd00009 | AAA | - | - | 206 | 366 | 1.16593E-17 |
| Coils | Coil | Coil | - | - | 437 | 457 | - |
| PANTHER | PTHR11638 | ATP-DEPENDENT CLP PROTEASE | - | - | 16 | 853 | 0.0 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.