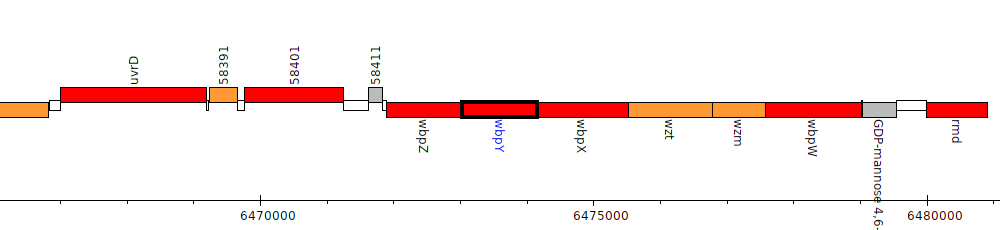
Pseudomonas aeruginosa LESB58, PALES_58431 (wbpY)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009103 | lipopolysaccharide biosynthetic process |
Inferred from Sequence or Structural Similarity
Term mapped from: PseudoCAP:PA5448
|
ECO:0000249 sequence similarity evidence used in automatic assertion |
9680202 | |
| Molecular Function | GO:0016757 | transferase activity, transferring glycosyl groups |
Inferred from Sequence Model
Term mapped from: InterPro:PF00534
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Lipopolysaccharide biosynthesis |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.50.2000 | Glycogen Phosphorylase B; | - | - | 186 | 356 | 1.2E-36 |
| CDD | cd03809 | GT4_MtfB-like | - | - | 2 | 367 | 8.8494E-112 |
| PANTHER | PTHR46401 | GLYCOSYLTRANSFERASE WBBK-RELATED | - | - | 1 | 371 | 1.3E-59 |
| Gene3D | G3DSA:3.40.50.2000 | Glycogen Phosphorylase B; | - | - | 2 | 185 | 1.9E-8 |
| SUPERFAMILY | SSF53756 | UDP-Glycosyltransferase/glycogen phosphorylase | - | - | 11 | 373 | 3.1E-66 |
| FunFam | G3DSA:3.40.50.2000:FF:000119 | Glycosyl transferase group 1 | - | - | 185 | 372 | 8.8E-69 |
| Pfam | PF00534 | Glycosyl transferases group 1 | IPR001296 | Glycosyl transferase, family 1 | 198 | 335 | 7.5E-26 |
| Pfam | PF13439 | Glycosyltransferase Family 4 | IPR028098 | Glycosyltransferase subfamily 4-like, N-terminal domain | 16 | 171 | 5.5E-9 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.