Pseudomonas aeruginosa RP73, M062_02215
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
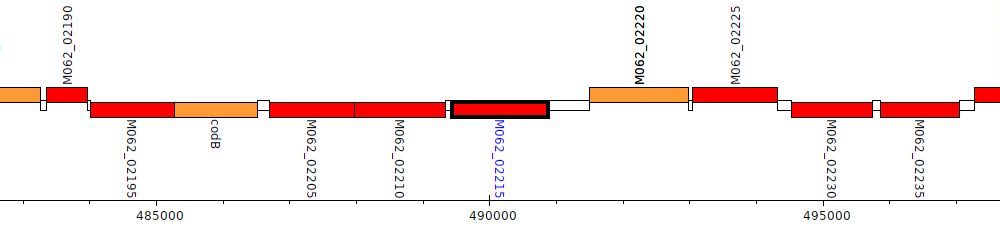
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa RP73
GCF_000414035.1|latest |
| Locus Tag |
M062_02215
|
| Name |
|
| Replicon | chromosome |
| Genomic location | 489435 - 490874 (- strand) |
Cross-References
| RefSeq | YP_008130907.1 |
| GI | 514408042 |
| Entrez | 15958186 |
| NCBI Locus Tag | M062_02215 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
phenylhydantoinase
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | -9.36 |
| Kyte-Doolittle Hydrophobicity Value | -0.171 |
| Molecular Weight (kDa) | 52.2 |
| Isoelectric Point (pI) | 6.17 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
AlphaFold 2 Protein Structure Predictions
Protein structure predictions using a neural network model developed by DeepMind. If a UniProtKB accession is associated with this protein, a search link will be provided below.
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 5E5C | HYDROLASE | 10/08/15 | Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1 | Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 / 1C / PRS 101 / LMG 12228) | 2.1 | X-RAY DIFFRACTION | 100.0 |
| 6KLK | HYDROLASE | 07/30/19 | Crystal structure of the Pseudomonas aeruginosa dihydropyrimidinase complexed with 5-FU | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 1.759 | X-RAY DIFFRACTION | 100.0 |
| 7E3U | HYDROLASE | 02/09/21 | Crystal structure of the Pseudomonas aeruginosa dihydropyrimidinase complexed with 5-AU | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 2.159 | X-RAY DIFFRACTION | 100.0 |
| 5YKD | HYDROLASE | 10/14/17 | Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1 at 2.17 angstrom resolution | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 2.17 | X-RAY DIFFRACTION | 100.0 |
| 6AJD | HYDROLASE | 08/27/18 | Crystal structure of a monometallic dihydropyrimidinase from Pseudomonas aeruginosa PAO1 reveals no lysine carbamylation within the active site | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 2.223 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 558 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000432 (819 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: M062_02215
Search term: phenylhydantoinase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
dihydropyrimidinase [Source:HGNC Symbol;Acc:HGNC:3013]
E-value:
9.8e-138
Percent Identity:
51.2
|
References
No references are associated with this feature.