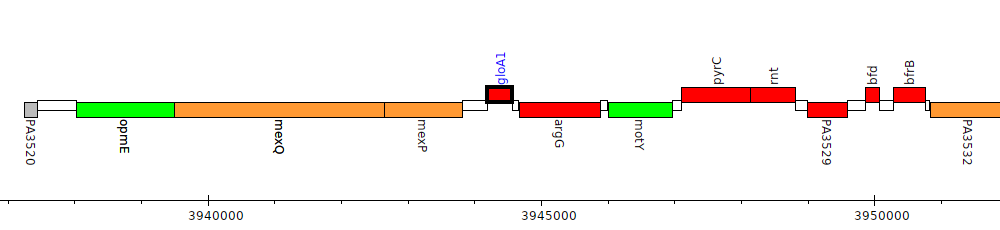
Pseudomonas aeruginosa PAO1, PA3524 (gloA1)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA3524
|
| Name |
gloA1
|
| Replicon | chromosome |
| Genomic location | 3944184 - 3944570 (+ strand) |
| Transposon Mutants | 2 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_252214.1 |
| GI | 15598720 |
| Affymetrix | PA3524_gloA1_at |
| DNASU | PaCD00007551 |
| Entrez | 879791 |
| GenBank | AAG06912.1 |
| INSDC | AAG06912.1 |
| NCBI Locus Tag | PA3524 |
| protein_id(GenBank) | gb|AAG06912.1|AE004773_1|gnl|PseudoCAP|PA3524 |
| TIGR | NTL03PA03524 |
| UniParc | UPI00000C5A53 |
| UniProtKB Acc | Q9HY85 |
| UniProtKB ID | Q9HY85_PSEAE |
| UniRef100 | UniRef100_Q9HY85 |
| UniRef50 | UniRef50_Q55595 |
| UniRef90 | UniRef90_Q9HY85 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
lactoylglutathione lyase
|
| Synonyms |
S-D-lactoylglutathione methylglyoxal lyase methylglyoxalase glyoxalase I ketone-aldehyde mutase aldoketomutase |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -6.08 |
| Kyte-Doolittle Hydrophobicity Value | -0.238 |
| Molecular Weight (kDa) | 14.3 |
| Isoelectric Point (pI) | 4.68 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 345 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG002141 (841 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA3524
Search term: gloA1
Search term: lactoylglutathione lyase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
glyoxalase I [Source:HGNC Symbol;Acc:HGNC:4323]
E-value:
1.1e-19
Percent Identity:
39.3
|
References
|
Overproduction and characterization of a dimeric non-zinc glyoxalase I from Escherichia coli: evidence for optimal activation by nickel ions.
Clugston SL, Barnard JF, Kinach R, Miedema D, Ruman R, Daub E, Honek JF
Biochemistry 1998 Jun 16;37(24):8754-63
PubMed ID: 9628737
|
|
Distinct classes of glyoxalase I: metal specificity of the Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis enzymes.
Sukdeo N, Clugston SL, Daub E, Honek JF
Biochem J 2004 Nov 15;384(Pt 1):111-7
PubMed ID: 15270717
|