Pseudomonas aeruginosa PAO1, PA3996 (lis)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
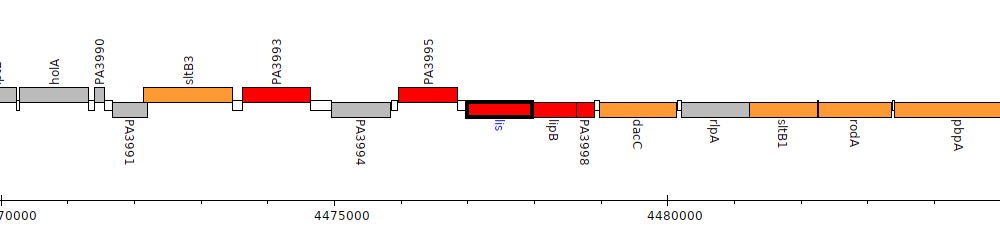
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA3996
|
| Name |
lis
Synonym: lipA |
| Replicon | chromosome |
| Genomic location | 4476994 - 4477977 (- strand) |
Cross-References
| RefSeq | NP_252685.1 |
| GI | 15599191 |
| Affymetrix | PA3996_lipA_at |
| DNASU | PaCD00007745 |
| Entrez | 878931 |
| GenBank | AAG07383.1 |
| INSDC | AAG07383.1 |
| NCBI Locus Tag | PA3996 |
| protein_id(GenBank) | gb|AAG07383.1|AE004817_7|gnl|PseudoCAP|PA3996 |
| TIGR | NTL03PA03996 |
| UniParc | UPI000012E6D0 |
| UniProtKB Acc | Q9HX25 |
| UniProtKB ID | LIPA_PSEAE |
| UniRef100 | UniRef100_B7V9C1 |
| UniRef50 | UniRef50_Q31F42 |
| UniRef90 | UniRef90_A6V0B4 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
lipoate synthase
|
| Synonyms |
lipoic acid synthetase |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -0.55 |
| Kyte-Doolittle Hydrophobicity Value | -0.398 |
| Molecular Weight (kDa) | 36.7 |
| Isoelectric Point (pI) | 6.90 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 495 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG001687 (536 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA3996
Search term: lis
Search term: lipoate synthase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
lipoic acid synthetase [Source:HGNC Symbol;Acc:HGNC:16429]
E-value:
1.2e-73
Percent Identity:
48.2
|
References
|
Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein.
Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, Broderick JB, Cronan JE Jr, Marletta MA
Biochemistry 2000 Dec 12;39(49):15166-78
PubMed ID: 11106496
|
|
Lipoic acid metabolism in Escherichia coli: isolation of null mutants defective in lipoic acid biosynthesis, molecular cloning and characterization of the E. coli lip locus, and identification of the lipoylated protein of the glycine cleavage system.
Vanden Boom TJ, Reed KE, Cronan JE Jr
J Bacteriol 1991 Oct;173(20):6411-20
PubMed ID: 1655709
|
|
The biosynthesis of lipoic acid. Cloning of lip, a lipoate biosynthetic locus of Escherichia coli.
Hayden MA, Huang I, Bussiere DE, Ashley GW
J Biol Chem 1992 May 15;267(14):9512-5
PubMed ID: 1577793
|
|
Lipoic acid metabolism in Escherichia coli: sequencing and functional characterization of the lipA and lipB genes.
Reed KE, Cronan JE Jr
J Bacteriol 1993 Mar;175(5):1325-36
PubMed ID: 8444795
|
|
Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein.
Morris TW, Reed KE, Cronan JE Jr
J Bacteriol 1995 Jan;177(1):1-10
PubMed ID: 8002607
|