Pseudomonas aeruginosa PAO1, PA4019 (ubiX)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
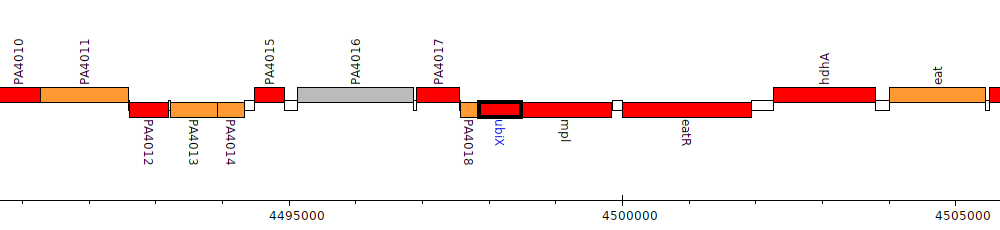
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA4019
|
| Name |
ubiX
|
| Replicon | chromosome |
| Genomic location | 4497850 - 4498479 (- strand) |
Cross-References
| RefSeq | NP_252708.1 |
| GI | 15599214 |
| Affymetrix | PA4019_at |
| DNASU | PaCD00007756 |
| Entrez | 878992 |
| GenBank | AAG07406.1 |
| INSDC | AAG07406.1 |
| NCBI Locus Tag | PA4019 |
| protein_id(GenBank) | gb|AAG07406.1|AE004818_12|gnl|PseudoCAP|PA4019 |
| TIGR | NTL03PA04019 |
| UniParc | UPI0000131194 |
| UniProtKB Acc | Q9HX08 |
| UniProtKB ID | PAAD_PSEAE |
| UniRef100 | UniRef100_Q9HX08 |
| UniRef50 | UniRef50_Q9HX08 |
| UniRef90 | UniRef90_Q9HX08 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
UbiX
|
| Synonyms |
probable aromatic acid decarboxylase |
| Evidence for Translation | |
| Charge (pH 7) | -6.20 |
| Kyte-Doolittle Hydrophobicity Value | 0.153 |
| Molecular Weight (kDa) | 22.4 |
| Isoelectric Point (pI) | 4.81 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 3ZQU | LYASE | 06/11/11 | STRUCTURE OF A PROBABLE AROMATIC ACID DECARBOXYLASE | PSEUDOMONAS AERUGINOSA | 1.5 | X-RAY DIFFRACTION | 100.0 |
| 6QLV | TRANSFERASE | 02/01/19 | Crystal structure of W200H UbiX in complex with a geranyl-FMN N5 adduct | Pseudomonas aeruginosa | 2.391 | X-RAY DIFFRACTION | 99.5 |
| 6QLJ | TRANSFERASE | 02/01/19 | Crystal structure of F181Q UbiX in complex with an oxidised N5-C1' adduct derived from DMAP | Pseudomonas aeruginosa | 1.77 | X-RAY DIFFRACTION | 99.5 |
| 4ZAW | LYASE | 04/14/15 | Structure of UbiX in complex with reduced prenylated FMN | Pseudomonas aeruginosa | 1.89 | X-RAY DIFFRACTION | 100.0 |
| 4ZAV | LYASE | 04/14/15 | UbiX in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | Pseudomonas aeruginosa | 1.4 | X-RAY DIFFRACTION | 100.0 |
| 4ZAY | LYASE | 04/14/15 | Structure of UbiX E49Q in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | Pseudomonas aeruginosa | 1.54 | X-RAY DIFFRACTION | 99.5 |
| 4ZAZ | LYASE | 04/14/15 | Structure of UbiX Y169F in complex with a covalent adduct formed between reduced FMN and dimethylallyl monophosphate | Pseudomonas aeruginosa | 1.45 | X-RAY DIFFRACTION | 99.5 |
| 4ZAX | TRANSFERASE | 04/14/15 | Structure of UbiX in complex with oxidised prenylated FMN (radical) | Pseudomonas aeruginosa | 1.61 | X-RAY DIFFRACTION | 100.0 |
| 6QLL | TRANSFERASE | 02/01/19 | Crystal structure of F181H UbiX in complex with FMN and dimethylallyl monophosphate | Pseudomonas aeruginosa | 1.56 | X-RAY DIFFRACTION | 99.5 |
| 6QLI | TRANSFERASE | 02/01/19 | Crystal structure of F181Q UbiX in complex with FMN and dimethylallyl monophosphate | Pseudomonas aeruginosa | 1.99 | X-RAY DIFFRACTION | 99.5 |
| 4ZAL | LYASE | 04/13/15 | Structure of UbiX E49Q mutant in complex with reduced FMN and dimethylallyl monophosphate | Pseudomonas aeruginosa | 1.62 | X-RAY DIFFRACTION | 99.5 |
| 6QLK | TRANSFERASE | 02/01/19 | Crystal structure of F181H UbiX in complex with prFMN | Pseudomonas aeruginosa | 1.78 | X-RAY DIFFRACTION | 99.5 |
| 6QLH | TRANSFERASE | 02/01/19 | Crystal structure of UbiX in complex with reduced FMN and isopentyl monophosphate | Pseudomonas aeruginosa | 1.57 | X-RAY DIFFRACTION | 100.0 |
| 4ZAN | LYASE | 04/13/15 | Structure of UbiX Y169F in complex with oxidised FMN and dimethylallyl monophosphate | Pseudomonas aeruginosa | 1.76 | X-RAY DIFFRACTION | 99.5 |
| 4ZAG | LYASE | 04/13/15 | Structure of UbiX E49Q mutant in complex with oxidised FMN and dimethylallyl monophosphate | Pseudomonas aeruginosa | 1.68 | X-RAY DIFFRACTION | 99.5 |
| 4ZAF | LYASE | 04/13/15 | Structure of UbiX in complex with oxidised FMN and dimethylallyl monophosphate | Pseudomonas aeruginosa | 1.71 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 371 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG001664 (536 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA4019
Search term: ubiX
Search term: UbiX
|
Human Homologs
References
|
Structure of PA4019, a putative aromatic acid decarboxylase from Pseudomonas aeruginosa.
Kopec J, Schnell R, Schneider G
Acta Crystallogr Sect F Struct Biol Cryst Commun 2011 Oct 1;67(Pt 10):1184-8
PubMed ID: 22102023
|
|
UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
White MD, Payne KA, Fisher K, Marshall SA, Parker D, Rattray NJ, Trivedi DK, Goodacre R, Rigby SE, Scrutton NS, Hay S, Leys D
Nature 2015 Jun 25;522(7557):502-6
PubMed ID: 26083743
|