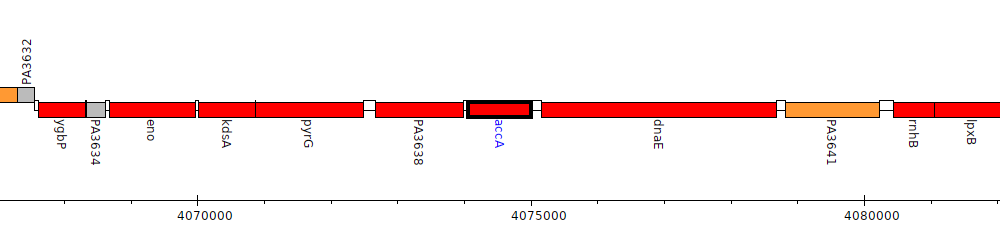
Pseudomonas aeruginosa PAO1, PA3639 (accA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0019541 | propionate metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0006090 | pyruvate metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0044255 | cellular lipid metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0003989 | acetyl-CoA carboxylase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR42853
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0009317 | acetyl-CoA carboxylase complex |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR42853
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016743 | carboxyl- or carbamoyltransferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR42853
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006633 | fatty acid biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR42853
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Fatty acid biosynthesis (path 1) |
ECO:0000037
not_recorded |
|||
| KEGG | pae00640 | Propanoate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01120 | Microbial metabolism in diverse environments | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00061 | Fatty acid biosynthesis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Propanoate metabolism |
ECO:0000037
not_recorded |
|||
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00620 | Pyruvate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01200 | Carbon metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01212 | Fatty acid metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Pyruvate metabolism |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.90.226.10 | - | - | - | 1 | 315 | 3.4E-106 |
| PRINTS | PR01069 | Acetyl-CoA carboxylase carboxyl transferase alpha subunit signature | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 169 | 182 | 2.2E-47 |
| PRINTS | PR01069 | Acetyl-CoA carboxylase carboxyl transferase alpha subunit signature | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 88 | 99 | 2.2E-47 |
| PANTHER | PTHR42853 | ACETYL-COENZYME A CARBOXYLASE CARBOXYL TRANSFERASE SUBUNIT ALPHA | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 3 | 314 | 0.0 |
| PRINTS | PR01069 | Acetyl-CoA carboxylase carboxyl transferase alpha subunit signature | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 134 | 150 | 2.2E-47 |
| NCBIfam | NF041504 | NCBIFAM: carboxyltransferase subunit alpha | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 58 | 311 | 2.4E-109 |
| Pfam | PF03255 | Acetyl co-enzyme A carboxylase carboxyltransferase alpha subunit | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 6 | 148 | 1.6E-63 |
| FunFam | G3DSA:3.90.226.10:FF:000008 | Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha | - | - | 4 | 316 | 0.0 |
| PRINTS | PR01069 | Acetyl-CoA carboxylase carboxyl transferase alpha subunit signature | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 113 | 131 | 2.2E-47 |
| NCBIfam | TIGR00513 | JCVI: acetyl-CoA carboxylase carboxyl transferase subunit alpha | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 2 | 315 | 0.0 |
| SUPERFAMILY | SSF52096 | ClpP/crotonase | IPR029045 | ClpP/crotonase-like domain superfamily | 5 | 314 | 4.42E-104 |
| PRINTS | PR01069 | Acetyl-CoA carboxylase carboxyl transferase alpha subunit signature | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 153 | 166 | 2.2E-47 |
| PRINTS | PR01069 | Acetyl-CoA carboxylase carboxyl transferase alpha subunit signature | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 197 | 206 | 2.2E-47 |
| Coils | Coil | Coil | - | - | 30 | 50 | - |
| Hamap | MF_00823 | Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha [accA]. | IPR001095 | Acetyl-CoA carboxylase, alpha subunit | 5 | 316 | 50.552128 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.