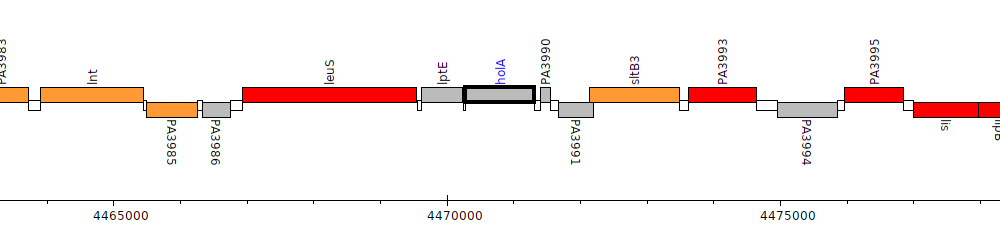
Pseudomonas aeruginosa PAO1, PA3989 (holA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006259 | DNA metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Cellular Component | GO:0009360 | DNA polymerase III complex | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
8505304 | Reviewed by curator |
| Molecular Function | GO:0003677 | DNA binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF06144
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006260 | DNA replication |
Inferred from Sequence Model
Term mapped from: InterPro:PF06144
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0009360 | DNA polymerase III complex |
Inferred from Sequence Model
Term mapped from: InterPro:PF06144
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003887 | DNA-directed DNA polymerase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF06144
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae03440 | Homologous recombination | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae03430 | Mismatch repair | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae03030 | DNA replication | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00240 | Pyrimidine metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00230 | Purine metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| FunFam | G3DSA:3.40.50.300:FF:001131 | DNA polymerase III subunit delta | - | - | 1 | 136 | 1.1E-68 |
| SUPERFAMILY | SSF48019 | post-AAA+ oligomerization domain-like | IPR008921 | DNA polymerase III, clamp loader complex, gamma/delta/delta subunit, C-terminal | 213 | 337 | 3.57E-32 |
| Gene3D | G3DSA:1.10.8.60 | - | - | - | 145 | 213 | 2.4E-27 |
| Pfam | PF06144 | DNA polymerase III, delta subunit | IPR010372 | DNA polymerase III delta, N-terminal | 20 | 191 | 4.5E-32 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 1 | 138 | 7.4E-47 |
| NCBIfam | TIGR01128 | JCVI: DNA polymerase III subunit delta | IPR005790 | DNA polymerase III, delta subunit | 16 | 336 | 1.4E-110 |
| FunFam | G3DSA:1.10.8.60:FF:000085 | DNA polymerase III subunit delta | - | - | 145 | 213 | 6.2E-37 |
| CDD | cd18138 | HLD_clamp_pol_III_delta | - | - | 143 | 207 | 9.62372E-27 |
| PANTHER | PTHR34388 | DNA POLYMERASE III SUBUNIT DELTA | - | - | 1 | 338 | 1.3E-98 |
| Pfam | PF14840 | Processivity clamp loader gamma complex DNA pol III C-term | IPR032780 | DNA polymerase III subunit delta, C-terminal | 216 | 339 | 8.6E-14 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 1 | 211 | 3.54E-49 |
| Gene3D | G3DSA:1.20.272.10 | - | - | - | 216 | 341 | 2.1E-36 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.