Pseudomonas aeruginosa UCBPP-PA14, PA14_18970 (pldA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
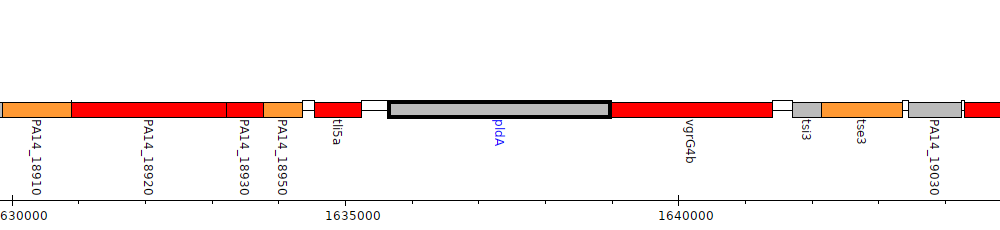
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa UCBPP-PA14 (Lee et al., 2006)
GCF_000014625.1|latest |
| Locus Tag |
PA14_18970
|
| Name |
pldA
Synonym: tle5 Synonym: type VI secretion lipase effector Tle5 |
| Replicon | chromosome |
| Genomic location | 1635662 - 1638973 (- strand) |
| Transposon Mutants | 2 transposon mutants in UCBPP-PA14 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
Cross-References
| RefSeq | YP_789660.1 |
| GI | 116051504 |
| Entrez | 4381470 |
| INSDC | ABJ12740.1 |
| NCBI Locus Tag | PA14_18970 |
| UniParc | UPI00003960CE |
| UniProtKB Acc | Q02QX6 |
| UniProtKB ID | Q02QX6_PSEAB |
| UniRef100 | UniRef100_Q02QX6 |
| UniRef50 | UniRef50_Q9HYC2 |
| UniRef90 | UniRef90_Q9HYC2 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
phospholipase D
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | -4.85 |
| Kyte-Doolittle Hydrophobicity Value | -0.312 |
| Molecular Weight (kDa) | 122.7 |
| Isoelectric Point (pI) | 6.60 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 7V53 | HYDROLASE | 08/16/21 | Crystal structure of full-length phospholipase D from Pseudomonas aeruginosa PAO1 | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 2.1 | X-RAY DIFFRACTION | 98.7 |
| 7WDK | IMMUNE SYSTEM | 12/21/21 | The structure of PldA-PA3488 complex | Pseudomonas aeruginosa; Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 3.05 | ELECTRON MICROSCOPY | 98.7 |
| 7V55 | HYDROLASE | 08/16/21 | Crystal structure of phospholipase D from Pseudomonas aeruginosa PAO1 using in situ proteolysis | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 3 | X-RAY DIFFRACTION | 98.7 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 18 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG002156 (79 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA14_18970
Search term: pldA
Search term: phospholipase D
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
phospholipase D1 [Source:HGNC Symbol;Acc:HGNC:9067]
E-value:
8.0e-26
Percent Identity:
30.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
phospholipase D1 [Source:HGNC Symbol;Acc:HGNC:9067]
E-value:
8.0e-26
Percent Identity:
30.7
|
References
|
Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model.
Wilderman PJ, Vasil AI, Johnson Z, Vasil ML
Mol Microbiol 2001 Jan;39(2):291-303
PubMed ID: 11136451
|
|
RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa.
Allsopp LP, Wood TE, Howard SA, Maggiorelli F, Nolan LM, Wettstadt S, Filloux A
Proc Natl Acad Sci U S A 2017 Jul 18;114(29):7707-7712
PubMed ID: 28673999
|
|
Biochemical characterization of a Pseudomonas aeruginosa phospholipase D.
Spencer C, Brown HA
Biochemistry 2015 Feb 10;54(5):1208-18
PubMed ID: 25565226
|