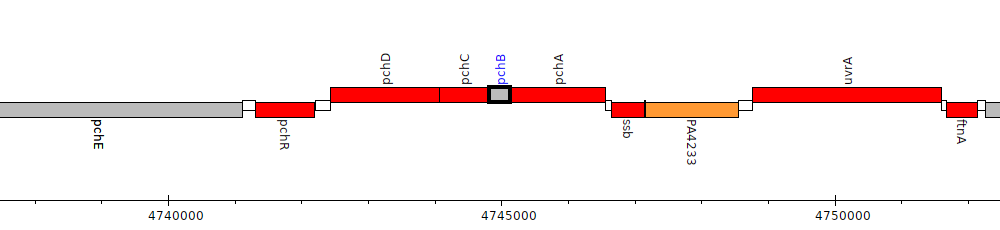
Pseudomonas aeruginosa PAO1, PA4230 (pchB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009697 | salicylic acid biosynthetic process | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
7500944 | Reviewed by curator |
| Biological Process | GO:0009697 | salicylic acid biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01803
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0046417 | chorismate metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:1.20.59.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016835 | carbon-oxygen lyase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01803
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004106 | chorismate mutase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01803
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae01053 | Biosynthesis of siderophore group nonribosomal peptides | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | PWY-6406 | salicylate biosynthesis I | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCAP | Pyochelin synthesis |
ECO:0000037
not_recorded |
|||
| KEGG | pae01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| NCBIfam | TIGR01803 | JCVI: chorismate mutase family protein | IPR008241 | Salicylate biosynthesis protein PchB | 10 | 90 | 5.5E-39 |
| SMART | SM00830 | CM_2_4 | IPR002701 | Chorismate mutase II, prokaryotic-type | 14 | 93 | 4.0E-23 |
| FunFam | G3DSA:1.20.59.10:FF:000012 | Isochorismate pyruvate lyase | - | - | 1 | 101 | 6.9E-70 |
| PIRSF | PIRSF029775 | Isochor_pyr_lyas | IPR008241 | Salicylate biosynthesis protein PchB | 1 | 101 | 1.5E-40 |
| Gene3D | G3DSA:1.20.59.10 | Chorismate mutase | IPR036979 | Chorismate mutase domain superfamily | 1 | 101 | 3.6E-27 |
| PANTHER | PTHR38041 | CHORISMATE MUTASE | - | - | 3 | 98 | 4.3E-22 |
| Pfam | PF01817 | Chorismate mutase type II | IPR002701 | Chorismate mutase II, prokaryotic-type | 14 | 90 | 1.9E-14 |
| SUPERFAMILY | SSF48600 | Chorismate mutase II | IPR036263 | Chorismate mutase type II superfamily | 1 | 94 | 5.36E-25 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.