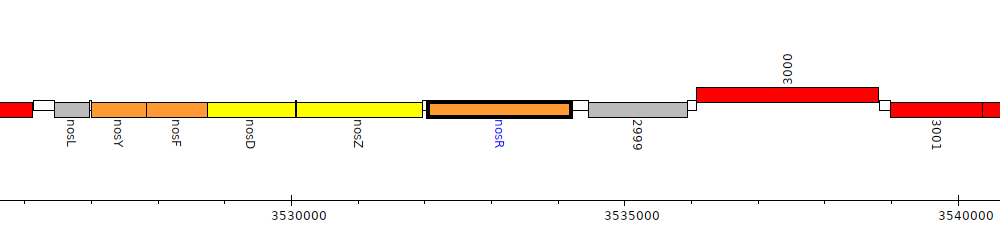
Pseudomonas fluorescens F113, PSF113_2998 (nosR)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0045893 | positive regulation of transcription, DNA-templated |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF036354
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003677 | DNA binding |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF036354
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0010181 | FMN binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF04205
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF036354
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF12801 | 4Fe-4S binding domain | IPR017896 | 4Fe-4S ferredoxin-type, iron-sulphur binding domain | 493 | 539 | 1.5E-15 |
| Pfam | PF12801 | 4Fe-4S binding domain | IPR017896 | 4Fe-4S ferredoxin-type, iron-sulphur binding domain | 594 | 640 | 1.1E-5 |
| SUPERFAMILY | SSF54862 | 4Fe-4S ferredoxins | - | - | 509 | 674 | 3.02E-7 |
| PANTHER | PTHR30224 | ELECTRON TRANSPORT PROTEIN | - | - | 70 | 705 | 0.0 |
| SMART | SM00900 | FMN_bind_2 | IPR007329 | FMN-binding | 79 | 172 | 2.1E-14 |
| Pfam | PF04205 | FMN-binding domain | IPR007329 | FMN-binding | 82 | 169 | 5.4E-6 |
| PIRSF | PIRSF036354 | NosR | IPR011399 | Nitrous oxide reductase expression regulator NosR | 2 | 716 | 0.0 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.