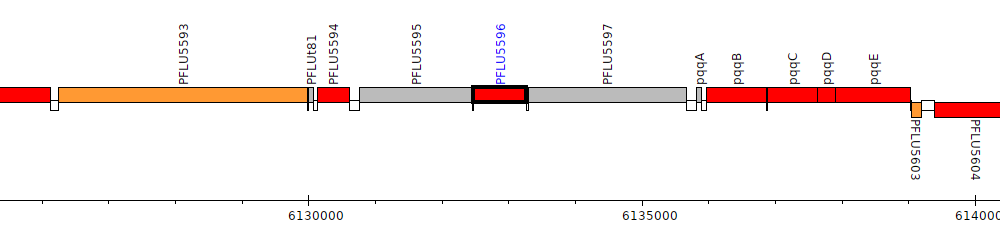
Pseudomonas fluorescens SBW25, PFLU5596
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006807 | nitrogen compound metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:PF00795
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016787 | hydrolase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd07576
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.60.110.10 | - | IPR036526 | Carbon-nitrogen hydrolase superfamily | 1 | 260 | 1.1E-76 |
| SUPERFAMILY | SSF56317 | Carbon-nitrogen hydrolase | IPR036526 | Carbon-nitrogen hydrolase superfamily | 1 | 261 | 3.66E-76 |
| Pfam | PF00795 | Carbon-nitrogen hydrolase | IPR003010 | Carbon-nitrogen hydrolase | 2 | 242 | 3.1E-55 |
| PANTHER | PTHR43674 | NITRILASE C965.09-RELATED | - | - | 2 | 261 | 2.4E-50 |
| CDD | cd07576 | R-amidase_like | IPR044083 | (R)-stereoselective amidase RamA-like | 2 | 256 | 4.49999E-130 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.