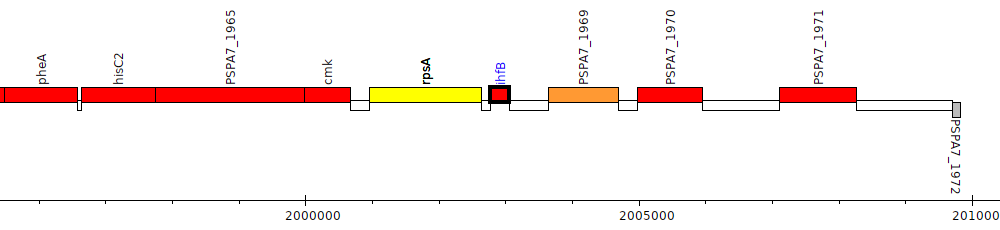
Pseudomonas aeruginosa PA7, PSPA7_1968 (ihfB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0030527 | structural constituent of chromatin |
Inferred from Sequence Model
Term mapped from: InterPro:SM00411
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006310 | DNA recombination |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00988
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003677 | DNA binding |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:4.10.520.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006355 | regulation of transcription, DNA-templated |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00988
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0005694 | chromosome |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00988
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR01727 | Prokaryotic integration host factor signature | IPR000119 | Histone-like DNA-binding protein | 59 | 72 | 6.8E-19 |
| NCBIfam | TIGR00988 | JCVI: integration host factor subunit beta | IPR005685 | Integration host factor, beta subunit | 1 | 94 | 7.2E-54 |
| Gene3D | G3DSA:4.10.520.10 | - | IPR010992 | Integration host factor (IHF)-like DNA-binding domain superfamily | 1 | 92 | 1.9E-30 |
| PRINTS | PR01727 | Prokaryotic integration host factor signature | IPR000119 | Histone-like DNA-binding protein | 41 | 56 | 6.8E-19 |
| FunFam | G3DSA:4.10.520.10:FF:000003 | Integration host factor subunit beta | - | - | 1 | 94 | 3.2E-50 |
| SMART | SM00411 | bhlneu | IPR000119 | Histone-like DNA-binding protein | 1 | 91 | 7.5E-45 |
| Pfam | PF00216 | Bacterial DNA-binding protein | IPR000119 | Histone-like DNA-binding protein | 1 | 91 | 9.4E-29 |
| CDD | cd13836 | IHF_B | - | - | 1 | 89 | 5.97196E-47 |
| PANTHER | PTHR33175 | DNA-BINDING PROTEIN HU | IPR000119 | Histone-like DNA-binding protein | 1 | 90 | 1.2E-27 |
| Hamap | MF_00381 | Integration host factor subunit beta [ihfB]. | IPR005685 | Integration host factor, beta subunit | 1 | 94 | 29.812973 |
| SUPERFAMILY | SSF47729 | IHF-like DNA-binding proteins | IPR010992 | Integration host factor (IHF)-like DNA-binding domain superfamily | 1 | 92 | 3.68E-31 |
| PRINTS | PR01727 | Prokaryotic integration host factor signature | IPR000119 | Histone-like DNA-binding protein | 75 | 89 | 6.8E-19 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.