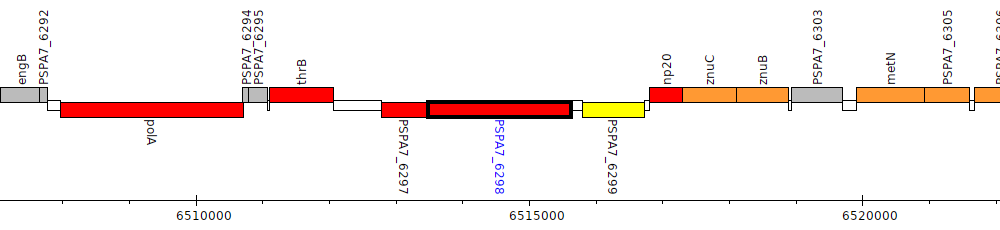
Pseudomonas aeruginosa PA7, PSPA7_6298
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0000166 | nucleotide binding |
Inferred from Sequence Model
Term mapped from: InterPro:cd02888
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0031419 | cobalamin binding |
Inferred from Sequence Model
Term mapped from: InterPro:cd02888
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor |
Inferred from Sequence Model
Term mapped from: InterPro:cd02888
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pap00230 | Purine metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pap01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pap00240 | Pyrimidine metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR43371 | VITAMIN B12-DEPENDENT RIBONUCLEOTIDE REDUCTASE | - | - | 10 | 705 | 0.0 |
| SUPERFAMILY | SSF51998 | PFL-like glycyl radical enzymes | - | - | 15 | 690 | 0.0 |
| CDD | cd02888 | RNR_II_dimer | IPR013344 | Ribonucleotide reductase, adenosylcobalamin-dependent | 75 | 651 | 0.0 |
| PRINTS | PR01183 | Ribonucleotide reductase large chain signature | IPR000788 | Ribonucleotide reductase large subunit, C-terminal | 316 | 327 | 2.2E-18 |
| PRINTS | PR01183 | Ribonucleotide reductase large chain signature | IPR000788 | Ribonucleotide reductase large subunit, C-terminal | 354 | 377 | 2.2E-18 |
| Gene3D | G3DSA:3.20.70.20 | - | - | - | 6 | 652 | 0.0 |
| FunFam | G3DSA:3.20.70.20:FF:000040 | Vitamin B12-dependent ribonucleotide reductase | - | - | 1 | 652 | 0.0 |
| PRINTS | PR01183 | Ribonucleotide reductase large chain signature | IPR000788 | Ribonucleotide reductase large subunit, C-terminal | 420 | 443 | 2.2E-18 |
| NCBIfam | TIGR02504 | JCVI: adenosylcobalamin-dependent ribonucleoside-diphosphate reductase | IPR013344 | Ribonucleotide reductase, adenosylcobalamin-dependent | 23 | 658 | 0.0 |
| Pfam | PF02867 | Ribonucleotide reductase, barrel domain | IPR000788 | Ribonucleotide reductase large subunit, C-terminal | 103 | 649 | 0.0 |
| PRINTS | PR01183 | Ribonucleotide reductase large chain signature | IPR000788 | Ribonucleotide reductase large subunit, C-terminal | 508 | 535 | 2.2E-18 |
| PRINTS | PR01183 | Ribonucleotide reductase large chain signature | IPR000788 | Ribonucleotide reductase large subunit, C-terminal | 392 | 414 | 2.2E-18 |
| PRINTS | PR01183 | Ribonucleotide reductase large chain signature | IPR000788 | Ribonucleotide reductase large subunit, C-terminal | 177 | 196 | 2.2E-18 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.