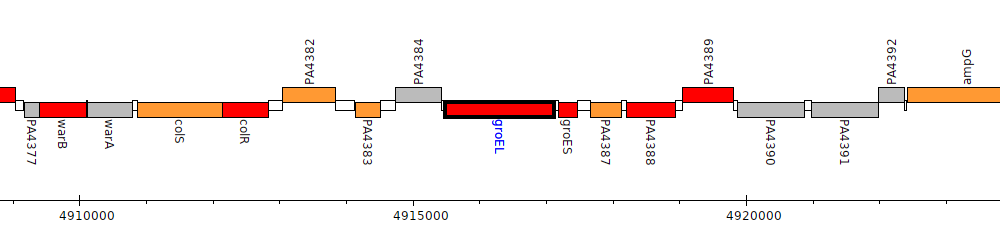
Pseudomonas aeruginosa PAO1, PA4385 (groEL)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00118
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0042026 | protein refolding |
Inferred from Sequence Model
Term mapped from: InterPro:PR00298
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0140662 | ATP-dependent protein folding chaperone |
Inferred from Sequence Model
Term mapped from: InterPro:PR00298
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae03018 | RNA degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR45633 | 60 KDA HEAT SHOCK PROTEIN, MITOCHONDRIAL | IPR001844 | Chaperonin Cpn60/GroEL | 2 | 530 | 0.0 |
| SUPERFAMILY | SSF52029 | GroEL apical domain-like | IPR027409 | GroEL-like apical domain superfamily | 184 | 376 | 2.62E-83 |
| Hamap | MF_00600 | Chaperonin GroEL [groEL]. | IPR001844 | Chaperonin Cpn60/GroEL | 2 | 546 | 41.565247 |
| FunFam | G3DSA:1.10.560.10:FF:000001 | 60 kDa chaperonin | - | - | 6 | 202 | 1.8E-68 |
| Gene3D | G3DSA:1.10.560.10 | - | IPR027413 | GroEL-like equatorial domain superfamily | 6 | 522 | 0.0 |
| Gene3D | G3DSA:3.50.7.10 | GroEL | IPR027409 | GroEL-like apical domain superfamily | 192 | 373 | 0.0 |
| PRINTS | PR00298 | 60kDa chaperonin signature | IPR001844 | Chaperonin Cpn60/GroEL | 83 | 110 | 7.4E-75 |
| NCBIfam | TIGR02348 | JCVI: chaperonin GroEL | IPR001844 | Chaperonin Cpn60/GroEL | 3 | 526 | 0.0 |
| Gene3D | G3DSA:3.30.260.10 | - | IPR027410 | TCP-1-like chaperonin intermediate domain superfamily | 137 | 410 | 0.0 |
| PRINTS | PR00298 | 60kDa chaperonin signature | IPR001844 | Chaperonin Cpn60/GroEL | 350 | 375 | 7.4E-75 |
| Pfam | PF00118 | TCP-1/cpn60 chaperonin family | IPR002423 | Chaperonin Cpn60/GroEL/TCP-1 family | 23 | 523 | 6.8E-91 |
| PRINTS | PR00298 | 60kDa chaperonin signature | IPR001844 | Chaperonin Cpn60/GroEL | 398 | 419 | 7.4E-75 |
| SUPERFAMILY | SSF54849 | GroEL-intermediate domain like | IPR027410 | TCP-1-like chaperonin intermediate domain superfamily | 137 | 203 | 3.43E-24 |
| SUPERFAMILY | SSF48592 | GroEL equatorial domain-like | IPR027413 | GroEL-like equatorial domain superfamily | 7 | 521 | 4.28E-75 |
| CDD | cd03344 | GroEL | IPR001844 | Chaperonin Cpn60/GroEL | 4 | 523 | 0.0 |
| PRINTS | PR00298 | 60kDa chaperonin signature | IPR001844 | Chaperonin Cpn60/GroEL | 27 | 53 | 7.4E-75 |
| FunFam | G3DSA:3.50.7.10:FF:000001 | 60 kDa chaperonin | - | - | 192 | 373 | 9.4E-89 |
| PRINTS | PR00298 | 60kDa chaperonin signature | IPR001844 | Chaperonin Cpn60/GroEL | 268 | 291 | 7.4E-75 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.