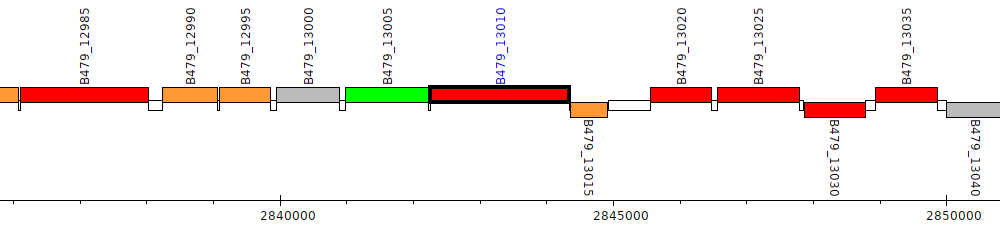
Pseudomonas putida HB3267, B479_13010
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas putida HB3267
GCF_000325725.1|latest |
| Locus Tag |
B479_13010
|
| Name |
|
| Replicon | chromosome |
| Genomic location | 2842257 - 2844341 (+ strand) |
Cross-References
| RefSeq | YP_007229441.1 |
| GI | 431802538 |
| Entrez | 14288989 |
| INSDC | AGA73498.1 |
| NCBI Locus Tag | B479_13010 |
| UniParc | UPI0002A15FA8 |
| UniProtKB Acc | L0FKH7 |
| UniProtKB ID | L0FKH7_PSEPU |
| UniRef100 | UniRef100_L0FKH7 |
| UniRef50 | UniRef50_L0FKH7 |
| UniRef90 | UniRef90_L0FKH7 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
3-hydroxybutyryl-CoA epimerase
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | -6.45 |
| Kyte-Doolittle Hydrophobicity Value | 0.011 |
| Molecular Weight (kDa) | 75567.1 |
| Isoelectric Point (pI) | 6.43 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 511 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG019905 (51 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: B479_13010
Search term: 3-hydroxybutyryl-CoA epimerase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:3247]
E-value:
5.9e-144
Percent Identity:
42.8
|
References
No references are associated with this feature.